Here we estimate the effects of the COVID-19 pandemic on Trust in Science at various stages of the pandemic in New Zealand.
Show code
# import data
df <- readRDS(here::here("data", "df"))
# prepare graph
ord_dates_class <- c(
"Baseline",
"JanFeb",
"EarlyMarch",
"Lockdown",
"PostLockdown")
# 3665 days after 30 June 2009 Saturday 13 July 2019
# 3842 days after 30 June 2009 Monday 6 January 2020
# 3895 days after 30 June 2009 Friday 28 February 2020
# 3921 days after 30 June 2009 WED MAR 25th
# 3954 days after 30 June 2009 Monday 27 April 2020
# 3672 days after June 30 2009 Saturday 20 July 2019
# prepare data
cv <- df %>%
dplyr::filter(YearMeasured == 1) %>%
dplyr::filter(Wave == 2019) %>%
droplevels() %>%
dplyr::mutate(Covid_Timeline =
as.factor(ifelse(
TSCORE %in% 3896:3921, # feb 29 - march 25th
"EarlyMarch",
ifelse(
TSCORE %in% 3922:3954,
"Lockdown", #march 26- Mon 27 April 2020
ifelse(
TSCORE > 3954, # after april 27th 20202
"PostLockdown",
ifelse(
TSCORE %in% 3842:3895, # jan 6 to feb 28
"JanFeb",
"Baseline" # 3672 TSCORE or 20 July 2019
)
)
)
) ))%>%
dplyr::select(
Wave,
TSCORE,
SCIENCE.TRUST,
Partner,
Employed,
Sat.Government,
Id,
Age,
Male,
Urban,
Edu,
Covid_Timeline,
EthnicCats,
Your.Personal.Relationships,
Urban
) %>%
dplyr::mutate(
)%>%
dplyr::mutate(Covid_Timeline = forcats::fct_relevel(Covid_Timeline, ord_dates_class)) %>%
droplevels() %>%
drop_na() %>%
mutate(SCIENCE.TRUST_S = scale(SCIENCE.TRUST),
Sat.Government_S = scale(Sat.Government),
Your.Personal.Relationships_S = scale(Your.Personal.Relationships))
The New Zealand Attitudes and Values Study
- Planned 20-year longitudinal study, currently in its 12\(^{th}\) year.
- Sample frame drawn randomly from NZ Electoral Roll.
- Postal questionnaire (coverage; retention ~ 80%)
- Focus on personality, social attitudes, values, religion, meaning-making, perfectionism, virtues, habits, employment, experiences of discrimination, physical and psychological health, and environmental attitudes …
- Current sample contains
36524unique people, which is \(>\) 1% of the NZ adult population. - Here, N = 38944 longitudinal participants who responded to both 2019/20 NZAVS waves.
Table of participants per condition
Show code
| Covid_Timeline | n |
|---|---|
| Baseline | 22093 |
| JanFeb | 2540 |
| EarlyMarch | 2574 |
| Lockdown | 3189 |
| PostLockdown | 8548 |
Method
Inverse Probability Weighting
To ensure balance across the condition in baseline covariates we use Inverse Probability Weighting.
Show code
#IPW data and check weights
cobalt::bal.tab(Covid_Timeline ~ Age + Edu + EthnicCats + Male + Urban,
data = cv, estimand = "ATE", m.threshold = .05)
Balance summary across all treatment pairs
Type Max.Diff.Un M.Threshold.Un
Age Contin. 0.3341 Not Balanced, >0.05
Edu_0 Binary 0.0086 Balanced, <0.05
Edu_1 Binary 0.0164 Balanced, <0.05
Edu_2 Binary 0.0032 Balanced, <0.05
Edu_3 Binary 0.0144 Balanced, <0.05
Edu_4 Binary 0.0121 Balanced, <0.05
Edu_5 Binary 0.0205 Balanced, <0.05
Edu_6 Binary 0.0094 Balanced, <0.05
Edu_7 Binary 0.0070 Balanced, <0.05
Edu_8 Binary 0.0046 Balanced, <0.05
Edu_9 Binary 0.0408 Balanced, <0.05
Edu_10 Binary 0.0116 Balanced, <0.05
EthnicCats_Euro Binary 0.0687 Not Balanced, >0.05
EthnicCats_Maori Binary 0.0534 Not Balanced, >0.05
EthnicCats_Pacific Binary 0.0131 Balanced, <0.05
EthnicCats_Asian Binary 0.0179 Balanced, <0.05
Male_Male Binary 0.0309 Balanced, <0.05
Urban_Urban Binary 0.0218 Balanced, <0.05
Balance tally for mean differences
count
Balanced, <0.05 15
Not Balanced, >0.05 3
Variable with the greatest mean difference
Variable Max.Diff.Un M.Threshold.Un
Age 0.3341 Not Balanced, >0.05
Sample sizes
Baseline JanFeb EarlyMarch Lockdown PostLockdown
All 22093 2540 2574 3189 8548Show code
# method by entropy balancing "which guarantees perfect balance on specified moments of the covariates while minimizing the entropy (a measure of dispersion) of the weights."
w_out <- WeightIt::weightit(Covid_Timeline ~ Age + Edu + EthnicCats + Male + Urban,
data = cv, estimand = "ATE", method = "ebal")
summary(w_out)
Summary of weights
- Weight ranges:
Min Max
Baseline 0.8366 |-----| 1.3358
JanFeb 0.7405 |------| 1.3377
EarlyMarch 0.3499 |-------------------------| 2.2038
Lockdown 0.5122 |-------------------------| 2.3630
PostLockdown 0.6712 |--------| 1.4220
- Units with 5 greatest weights by group:
12469 2083 5003 744 3950
Baseline 1.2609 1.2617 1.2777 1.2924 1.3358
6792 20019 18134 30943 14246
JanFeb 1.3215 1.3253 1.3275 1.3333 1.3377
3157 4943 581 3243 889
EarlyMarch 2.0088 2.0361 2.0451 2.0799 2.2038
16425 8141 8087 31514 5347
Lockdown 1.9601 2.1371 2.1874 2.2753 2.363
19492 31240 8945 33627 20012
PostLockdown 1.3766 1.3827 1.3943 1.3953 1.422
- Weight statistics:
Coef of Var MAD Entropy # Zeros
Baseline 0.060 0.044 0.002 0
JanFeb 0.092 0.074 0.004 0
EarlyMarch 0.264 0.214 0.035 0
Lockdown 0.214 0.163 0.022 0
PostLockdown 0.102 0.081 0.005 0
- Effective Sample Sizes:
Baseline JanFeb EarlyMarch Lockdown
Unweighted 22093. 2540. 2574. 3189.
Weighted 22015.01 2518.65 2406.51 3049.22
PostLockdown
Unweighted 8548.
Weighted 8460.07Inspect weights:
Show code
cobalt::bal.tab(w_out, m.threshold = .05, disp.v.ratio = TRUE)
Call
WeightIt::weightit(formula = Covid_Timeline ~ Age + Edu + EthnicCats +
Male + Urban, data = cv, method = "ebal", estimand = "ATE")
Balance summary across all treatment pairs
Type Max.Diff.Adj M.Threshold
Age Contin. 0.0002 Balanced, <0.05
Edu_0 Binary 0.0001 Balanced, <0.05
Edu_1 Binary 0.0000 Balanced, <0.05
Edu_2 Binary 0.0001 Balanced, <0.05
Edu_3 Binary 0.0001 Balanced, <0.05
Edu_4 Binary 0.0001 Balanced, <0.05
Edu_5 Binary 0.0001 Balanced, <0.05
Edu_6 Binary 0.0000 Balanced, <0.05
Edu_7 Binary 0.0002 Balanced, <0.05
Edu_8 Binary 0.0001 Balanced, <0.05
Edu_9 Binary 0.0000 Balanced, <0.05
Edu_10 Binary 0.0001 Balanced, <0.05
EthnicCats_Euro Binary 0.0000 Balanced, <0.05
EthnicCats_Maori Binary 0.0000 Balanced, <0.05
EthnicCats_Pacific Binary 0.0001 Balanced, <0.05
EthnicCats_Asian Binary 0.0000 Balanced, <0.05
Male_Male Binary 0.0001 Balanced, <0.05
Urban_Urban Binary 0.0000 Balanced, <0.05
Max.V.Ratio.Adj
Age 1.1722
Edu_0 .
Edu_1 .
Edu_2 .
Edu_3 .
Edu_4 .
Edu_5 .
Edu_6 .
Edu_7 .
Edu_8 .
Edu_9 .
Edu_10 .
EthnicCats_Euro .
EthnicCats_Maori .
EthnicCats_Pacific .
EthnicCats_Asian .
Male_Male .
Urban_Urban .
Balance tally for mean differences
count
Balanced, <0.05 18
Not Balanced, >0.05 0
Variable with the greatest mean difference
Variable Max.Diff.Adj M.Threshold
Edu_7 0.0002 Balanced, <0.05
Effective sample sizes
Baseline JanFeb EarlyMarch Lockdown
Unadjusted 22093. 2540. 2574. 3189.
Adjusted 22015.01 2518.65 2406.51 3049.22
PostLockdown
Unadjusted 8548.
Adjusted 8460.07Estimate model
Model
Show code
SCIENCE.TRUST <- glm(
SCIENCE.TRUST ~ Covid_Timeline,
data = cv19w,
weights = ipw
)
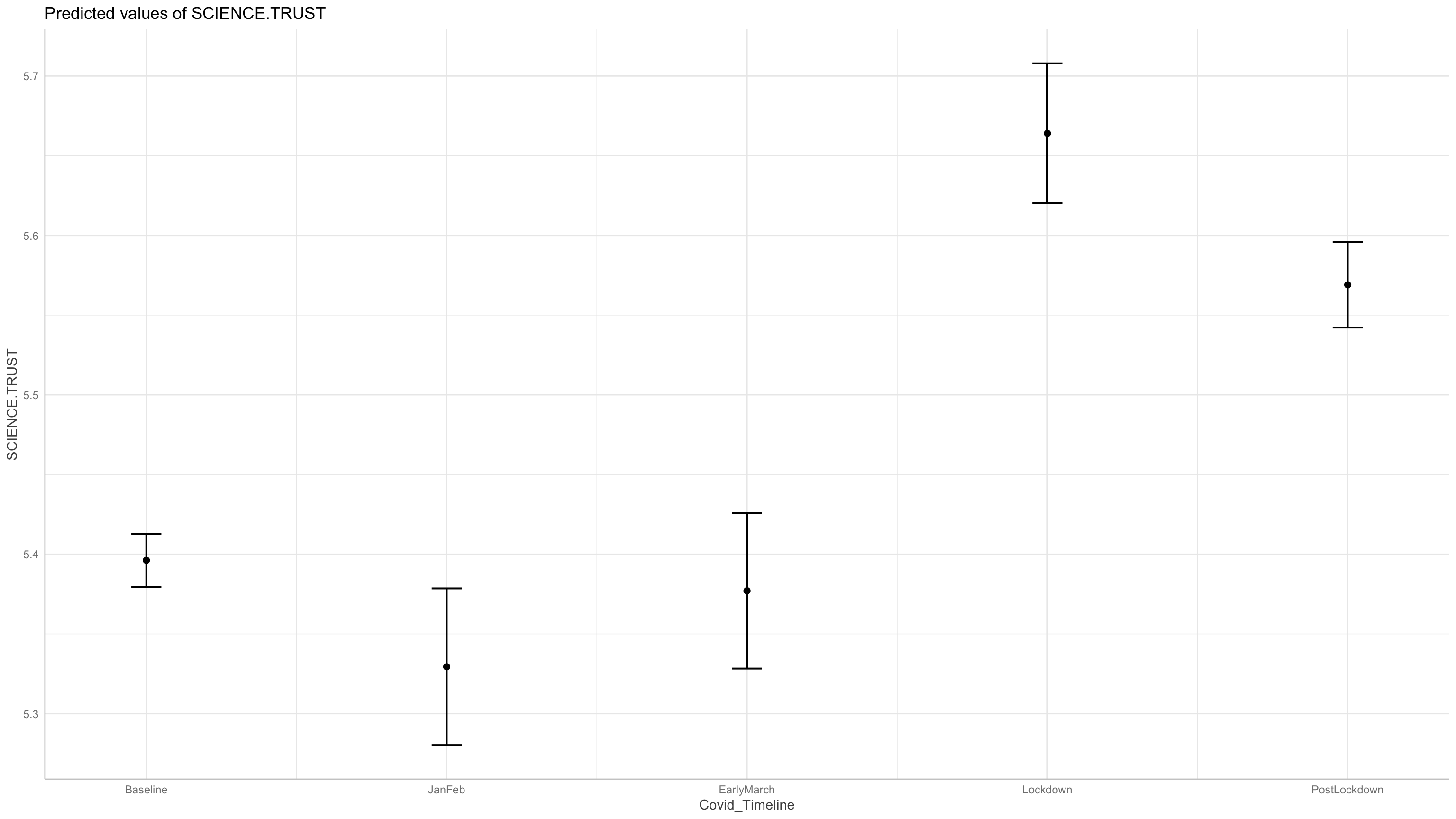
Marginal means
Show code
# estimated marginal means
means <- estimate_means(SCIENCE.TRUST, at = "Covid_Timeline")
# print
means%>%
print_md()
| Covid_Timeline | Mean | SE | CI_low | CI_high |
|---|---|---|---|---|
| Baseline | 5.40 | 8.50e-03 | 5.38 | 5.41 |
| JanFeb | 5.33 | 0.03 | 5.28 | 5.38 |
| EarlyMarch | 5.38 | 0.02 | 5.33 | 5.43 |
| Lockdown | 5.66 | 0.02 | 5.62 | 5.71 |
| PostLockdown | 5.57 | 0.01 | 5.54 | 5.60 |
Marginal means estimated at Covid_Timeline
Show code
#
contrasts <- estimate_contrasts(SCIENCE.TRUST, contrast = "Covid_Timeline", adjust = "bonferroni")
contrasts%>%
print_md()
| Level1 | Level2 | Difference | CI_low | CI_high | SE | df | t | p |
|---|---|---|---|---|---|---|---|---|
| Baseline | EarlyMarch | 0.02 | -0.05 | 0.09 | 0.03 | 38939 | 0.73 | 0.95 |
| Baseline | JanFeb | 0.07 | -7.54e-03 | 0.14 | 0.03 | 38939 | 2.52 | 0.09 |
| Baseline | Lockdown | -0.27 | -0.34 | -0.20 | 0.02 | 38939 | -11.19 | 1.33e-14 |
| Baseline | PostLockdown | -0.17 | -0.22 | -0.13 | 0.02 | 38939 | -10.73 | 4.87e-14 |
| EarlyMarch | Lockdown | -0.29 | -0.38 | -0.19 | 0.03 | 38939 | -8.57 | 4.54e-14 |
| EarlyMarch | PostLockdown | -0.19 | -0.27 | -0.11 | 0.03 | 38939 | -6.75 | 1.45e-10 |
| JanFeb | EarlyMarch | -0.05 | -0.15 | 0.05 | 0.04 | 38939 | -1.35 | 0.66 |
| JanFeb | Lockdown | -0.33 | -0.43 | -0.24 | 0.03 | 38939 | -9.96 | 4.20e-14 |
| JanFeb | PostLockdown | -0.24 | -0.32 | -0.16 | 0.03 | 38939 | -8.39 | 5.16e-14 |
| Lockdown | PostLockdown | 0.10 | 0.02 | 0.17 | 0.03 | 38939 | 3.62 | 2.68e-03 |
Marginal contrasts estimated at Covid_Timeline p-value adjustment method: Bonferroni
Graph of the expected (marginal) means:
Show code
#plot(means, alpha =.005)
ggplot(cv19w, aes(x = Covid_Timeline, y = SCIENCE.TRUST)) +
# Add base data
geom_violin(aes(fill = Covid_Timeline), color = "white") +
geom_jitter2(width = 0.05, alpha = 0.005) +
# Add pointrange and line from means
geom_line(data = means, aes(y = Mean, group = 1), size = .3) +
geom_pointrange(
data = means,
aes(y = Mean, ymin = CI_low, ymax = CI_high),
size = .3,
color = "white"
) +
# Improve colors
scale_fill_material() +
theme_modern() +
scale_color_discrete()
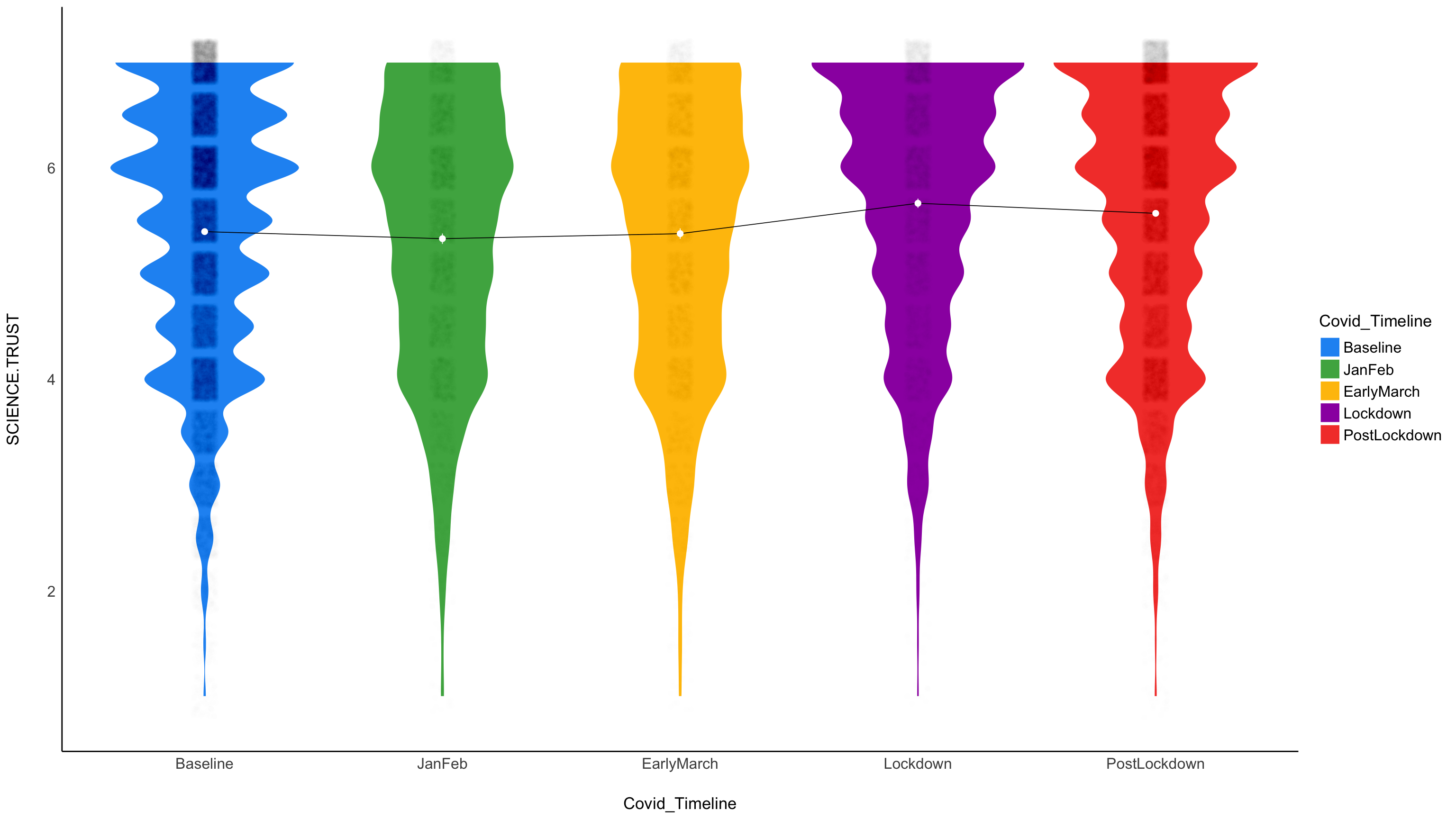
Trust in science increased, and then dropped. The key question, will it trust revert to baseline (or worse)?
Below another graph of the same result.
Causal mediation model
Show code
library(mediation)
#length(unique(cv19n$Id))
# + Age + Edu + EthnicCats + Male + Relid + Partner + Urban
med.fit <- lm( Sat.Government_S ~ Covid_Timeline + Age + Edu + EthnicCats + Male + Urban, data = cv19w)
out.fit <- lm( SCIENCE.TRUST_S ~ Sat.Government_S + Covid_Timeline + Age + Edu + EthnicCats + Male + Urban,
data = cv19w)
med.out <- mediate(med.fit, out.fit, treat = "Covid_Timeline", mediator = "Sat.Government_S",
robustSE = TRUE, sims = 1000, control.value = "Baseline", treat.value = "Lockdown")
# save model
saveRDS(med.out, here::here("_posts", "covscience","mods", "med.out.rds"))
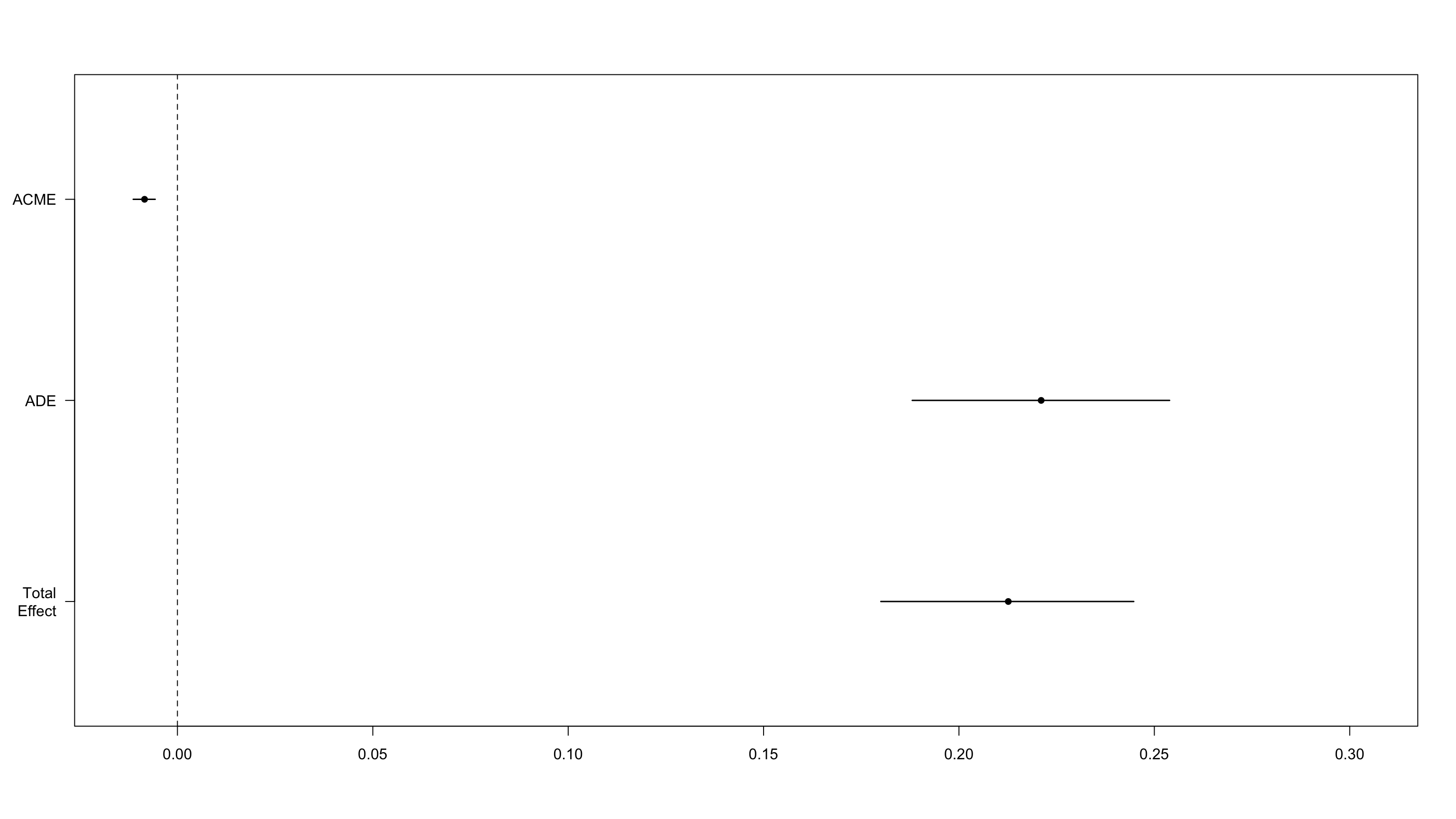
We observe that government satisfaction strongly mediates the effect of Lockdown on trust in science.
Show code
Causal Mediation Analysis
Quasi-Bayesian Confidence Intervals
Estimate 95% CI Lower 95% CI Upper p-value
ACME 0.1041 0.0951 0.11 <2e-16
ADE 0.1078 0.0726 0.14 <2e-16
Total Effect 0.2119 0.1775 0.25 <2e-16
Prop. Mediated 0.4923 0.4184 0.60 <2e-16
ACME ***
ADE ***
Total Effect ***
Prop. Mediated ***
---
Signif. codes:
0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Sample Size Used: 38944
Simulations: 1000 We next contrast the Locdown with the period following lockdown in which there was declining trust in science.
Sensitivity Analysis
Sensitivity plot reveals that for sequential ignorability to be violated, the correlation between the the error terms of the mediator and outcome (rho) would need to be .25. We infer that the findings are reasonably robust in respect of unmeasured mediator outcome confounding.
Show code
sens.out <- readRDS(sens.out, here::here("_posts", "covscience","mods", "sens.out.rds"))
#summary(sens.out)
plot(sens.out)
#plot(sens.out, sens.par = "R2", r.type = "total", sign.prod = "positive")
# This is the workhorse function for sensitivity analyses for average causal mediation effects. The sensitivity analysis can be used to assess the robustness of the findings from mediate to the violation of sequential ignorability, the crucial identification assumption necessary for the estimates to be valid. The analysis proceeds by quantifying the degree of sequential ignorability violation as the correlation between the error terms of the mediator and outcome models, and then calculating the true values of the average causal mediation effect for given values of this sensitivity parameter, rho. The original findings are deemed sensitive if the true effects are found to vary widely as function of rho.
#The resulting "rho" figures plot the estimated true values of ACME (or ADE, proportion mediated) against rho, along with the confidence intervals. When rho is zero, sequantial ignorability holds, so the estimated value at that point will be equal to the estimate returned by the mediate. The confidence level is determined by the 'conf.level' value of the original mediate object
Negative Controls
Show code
med.fitR <- lm( Your.Personal.Relationships_S ~ Covid_Timeline + Age + Edu + EthnicCats + Male + Urban, data = cv19w)
out.fitR <- lm( SCIENCE.TRUST_S ~ Your.Personal.Relationships_S + Covid_Timeline + Age + Edu + EthnicCats + Male + Urban,
data = cv19w)
med.outR <- mediate(med.fitR, out.fitR, treat = "Covid_Timeline", mediator = "Your.Personal.Relationships_S",
robustSE = TRUE, sims = 1000, control.value = "Baseline", treat.value = "Lockdown")
# save model
saveRDS(med.outR, here::here("_posts", "covscience","mods", "med.outR.rds"))
We do not find an effect for satisfaction with relationships
Show code
Causal Mediation Analysis
Quasi-Bayesian Confidence Intervals
Estimate 95% CI Lower 95% CI Upper p-value
ACME -0.00841 -0.01132 -0.01 <2e-16
ADE 0.22104 0.18804 0.25 <2e-16
Total Effect 0.21262 0.18001 0.24 <2e-16
Prop. Mediated -0.03920 -0.05643 -0.03 <2e-16
ACME ***
ADE ***
Total Effect ***
Prop. Mediated ***
---
Signif. codes:
0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Sample Size Used: 38944
Simulations: 1000 Show code
plot(med.outR)
Reverse causation
We next investigate whether declining satisfaction with government caused a drop in trust in science (reverse causal model)
We find evidence for only a weak mediation effect
Show code
Causal Mediation Analysis
Quasi-Bayesian Confidence Intervals
Estimate 95% CI Lower 95% CI Upper p-value
ACME -0.0222 -0.0288 -0.02 <2e-16
ADE -0.0542 -0.0881 -0.02 <2e-16
Total Effect -0.0765 -0.1104 -0.04 <2e-16
Prop. Mediated 0.2863 0.1853 0.60 <2e-16
ACME ***
ADE ***
Total Effect ***
Prop. Mediated ***
---
Signif. codes:
0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Sample Size Used: 39746
Simulations: 1000 Show code
plot(med.out2)
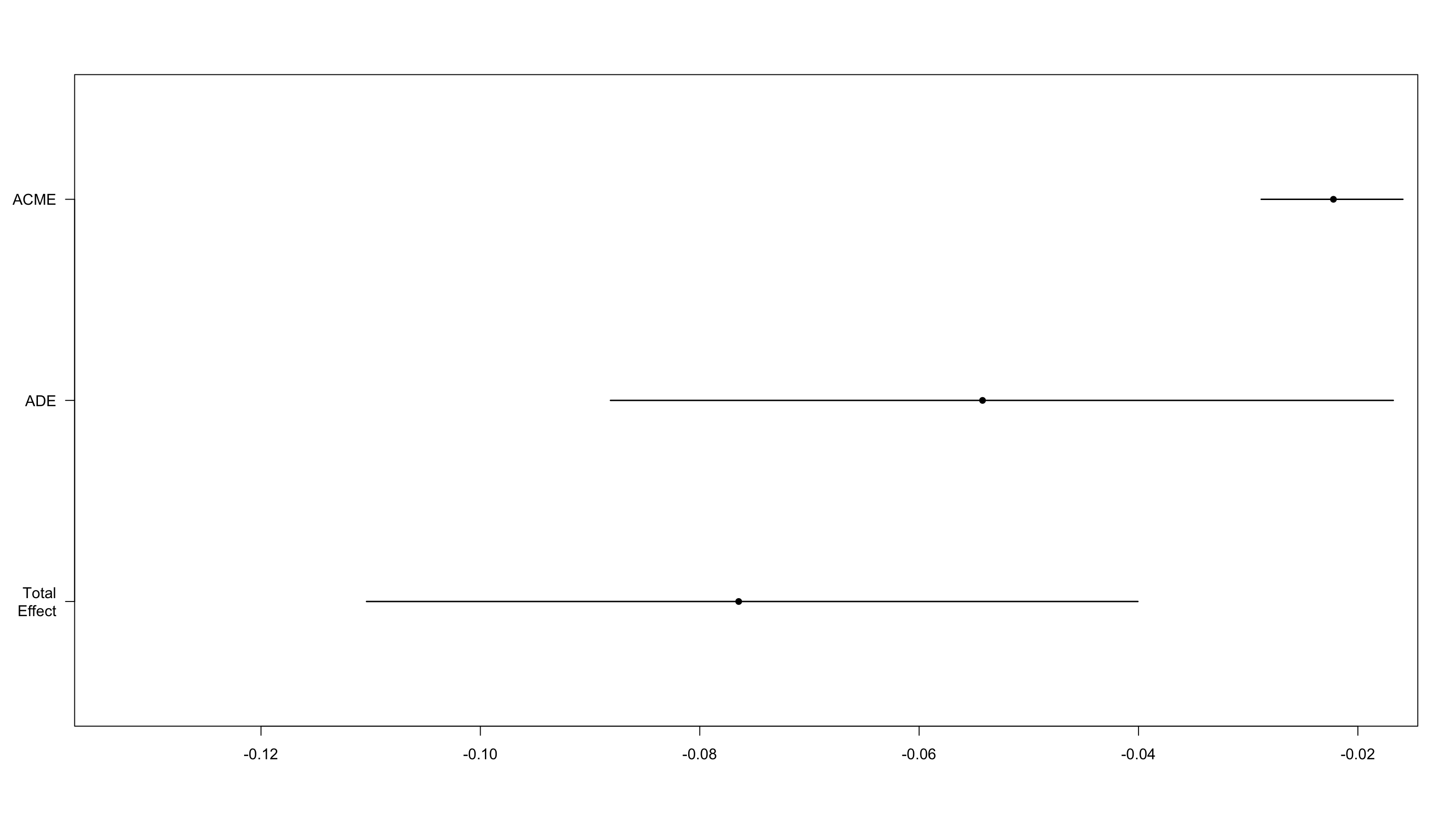
As expected this small effect is robust to unmeasured confounding.
Does trust in science mediate the effect of government satisfaction?
Show code
med.fitb <- lm( SCIENCE.TRUST_S ~ Covid_Timeline + Age + Edu + EthnicCats + Male + Urban, data = cv19w)
out.fitb <- lm( Sat.Government_S ~ SCIENCE.TRUST_S + Covid_Timeline + Age + Edu + EthnicCats + Male + Urban,
data = cv19w)
med.outb <- mediate(med.fitb, out.fitb, treat = "Covid_Timeline", mediator = "SCIENCE.TRUST_S",
robustSE = TRUE, sims = 1000, control.value = "Baseline", treat.value = "Lockdown")
saveRDS(med.outb, here::here("_posts", "covscience","mods", "med.outb.rds"))
We find a very strong effect of lockdown on satisfaction with government, but we do not find any evidence for a substantial effect of mediation through trust in science.
Show code
Causal Mediation Analysis
Quasi-Bayesian Confidence Intervals
Estimate 95% CI Lower 95% CI Upper p-value
ACME 0.0362 0.0299 0.04 <2e-16
ADE 0.5815 0.5472 0.62 <2e-16
Total Effect 0.6177 0.5850 0.65 <2e-16
Prop. Mediated 0.0584 0.0485 0.07 <2e-16
ACME ***
ADE ***
Total Effect ***
Prop. Mediated ***
---
Signif. codes:
0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Sample Size Used: 39746
Simulations: 1000 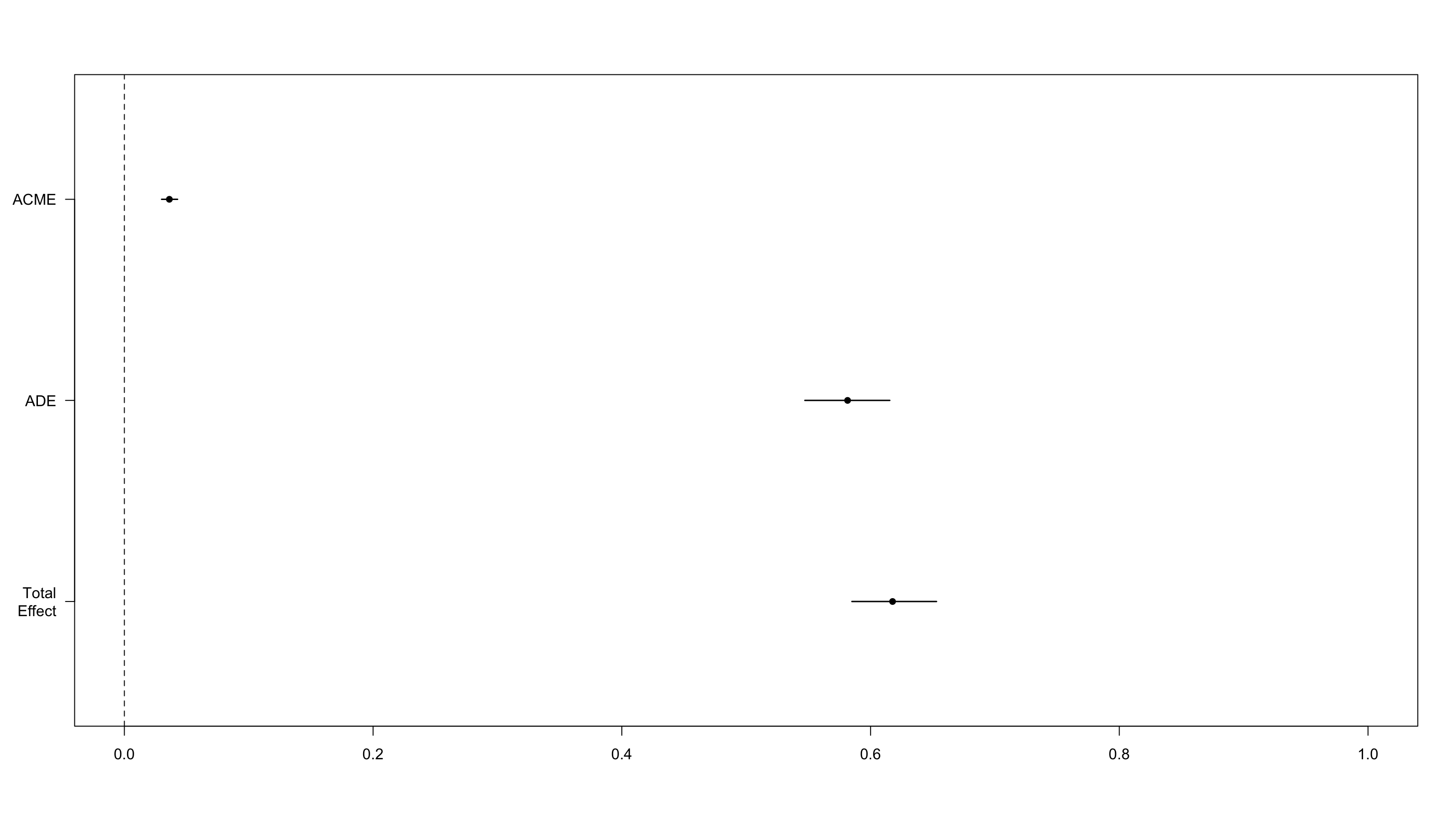
Sensitivity analysis
Show code
sens.outb2 <- medsens(med.outb2, rho.by = 0.25, effect.type = "indirect", sims = 100)
# save sensitivity analysis
saveRDS(sens.outb2, here::here("_posts", "covscience","mods", "sens.outb2.rds"))
sens.outb2 <- readRDS(here::here("_posts", "covscience","mods", "sens.outb2.rds"))
plot(sens.outb2)
summary(sens.outb2)
Loss of gov’t satisfaction does not mediate loss of trust in science.
We can ask whether the fall in government satisfaction was mediated by a loss of confidence in science.
We do not find strong evidence for a causal effect of loss of trust in science on satisfaction with the government.
Show code
Causal Mediation Analysis
Quasi-Bayesian Confidence Intervals
Estimate 95% CI Lower 95% CI Upper p-value
ACME -0.0130 -0.0194 -0.01 <2e-16
ADE -0.1197 -0.1575 -0.08 <2e-16
Total Effect -0.1326 -0.1715 -0.09 <2e-16
Prop. Mediated 0.0973 0.0497 0.16 <2e-16
ACME ***
ADE ***
Total Effect ***
Prop. Mediated ***
---
Signif. codes:
0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Sample Size Used: 39746
Simulations: 1000 Show code
# covid timeline
# 3665 days after 30 June 2009 Saturday 13 July 2019
# 3842 days after 30 June 2009 Monday 6 January 2020
# 3895 days after 30 June 2009 Friday 28 February 2020
# 3921 days after 30 June 2009 WED MAR 25th
library(lubridate)
library(tidyverse)
carep <- cv %>%
dplyr::mutate(timeline = make_date(year = 2009, month = 6, day = 30) + TSCORE) %>%
count(day = floor_date(timeline, "day")) %>%
dplyr::mutate(condition = ifelse(
# day >= "2019-06-20" &
day < "2020-01-06",
"Baseline",
ifelse(
day >= "2020-01-06" & day < "2020-02-29",
"JanFeb",
ifelse(
day >= "2020-02-29" & day < "2020-03-25",
"EarlyMarch",
ifelse(
day >= "2020-03-25" &
day < "2020-04-28",
"Lockdown",
"PostLockdown"
))))) %>%
dplyr::mutate(condition = forcats::fct_relevel(condition, ord_dates_class)) %>%
arrange(day)
# get COVID date
minday <- min(carep$day) # min day
dates_vline1<- as.Date(minday) # min day
dates_vline2<- as.Date("2020-01-06")
dates_vline3<- as.Date("2020-02-28")
# get data of change of baseline
dates_vline4<- as.Date("2020-03-25")
dates_vline5 <- as.Date("2020-04-27")
dates_vline6 <- as.Date("2020-10-15") # end day
# for line in a graph
# for line in a graph
#
dates_vline1b <- which(carep$day %in% dates_vline1)
dates_vline2b <- which(carep$day %in% dates_vline2)
dates_vline3b <- which(carep$day %in% dates_vline3)
dates_vline4b <- which(carep$day %in% dates_vline4)
dates_vline5b <- which(carep$day %in% dates_vline5)
dates_vline6b <- which(carep$day %in% dates_vline6)
tline <- ggplot(carep, aes(day, n)) +
geom_col(aes(fill = condition)) + scale_x_date(date_labels = "%b/%Y", limits = c(as.Date(minday),
as.Date("2020-10-15"))) +
scale_y_continuous(limits = c(0, 1000)) +
geom_vline(xintercept = as.numeric(carep$day[dates_vline1b]),
col = "red",
linetype = "dashed") +
geom_vline(xintercept = as.numeric(carep$day[dates_vline2b]),
col = "red") +
geom_vline(xintercept = as.numeric(carep$day[dates_vline3b]),
col = "red",
linetype = "dashed") +
geom_vline(xintercept = as.numeric(carep$day[dates_vline4b]),
col = "red",
linetype = "dashed") +
geom_vline(xintercept = as.numeric(carep$day[dates_vline5b]),
col = "red",
linetype = "dashed") +
xlab("NZAVS Waves 10 and 11 daily counts: repeated measures") + ylab("Count of Responses") + theme_classic() + scale_fill_viridis_d() + theme(
legend.position = "top",
legend.text = element_text(size = 6),
legend.title = element_text(color = "Black", size = 8)) +
# annotate(
# "rect",
# xmin = as.Date("2019-06-20"),
# xmax = dates_vline1,
# ymin = 0,
# ymax = 1000,
# alpha = .1,
# fill = "darkblue"
# ) +
# annotate(
# "rect",
# xmin = dates_vline1,
# xmax = dates_vline6,
# ymin = 0,
# ymax = 1000,
# alpha = .001
# ) +
# annotate(
# geom = "curve",
# x = as.Date("2019-06-20"),
# y = 750,
# xend = as.Date("2019-10-15"),
# yend = 800,
# curvature = -.2,
# arrow = arrow(length = unit(2, "mm"))
# ) +
# annotate(
# geom = "curve",
# x = as.Date("2019-06-20"),
# y = 750,
# xend = as.Date("2020-01-30"),
# yend = 850,
# curvature = -.2,
# arrow = arrow(length = unit(2, "mm"))
# ) +
# annotate(
# geom = "curve",
# x = as.Date("2019-06-20"),
# y = 750,
# xend = as.Date("2020-03-10"),
# yend = 875,
# curvature = -.2,
# arrow = arrow(length = unit(2, "mm"))
# ) +
# annotate(
# geom = "curve",
# x = as.Date("2019-06-20"),
# y = 750,
# xend = as.Date("2020-04-10"),
# yend = 900,
# curvature = -.2,
# arrow = arrow(length = unit(2, "mm"))
# ) +
# annotate(
# geom = "curve",
# x = as.Date("2019-06-20"),
# y = 750,
# xend = as.Date("2020-06-10"),
# yend = 925,
# curvature = -.2,
# arrow = arrow(length = unit(2, "mm"))
# ) +
annotate("text",
x = as.Date("2019-06-20"),
y = 755,
label = "0") +
annotate("text",
x = as.Date("2019-10-25"),
y = 800,
label = "1") +
annotate("text",
x = as.Date("2020-02-10"),
y = 850,
label = "2") +
annotate("text",
x = as.Date("2020-03-15"),
y = 875,
label = "3") +
annotate("text",
x = as.Date("2020-04-20"),
y = 900,
label = "4") +
annotate("text",
x = as.Date("2020-06-10"),
y = 925,
label = "5")
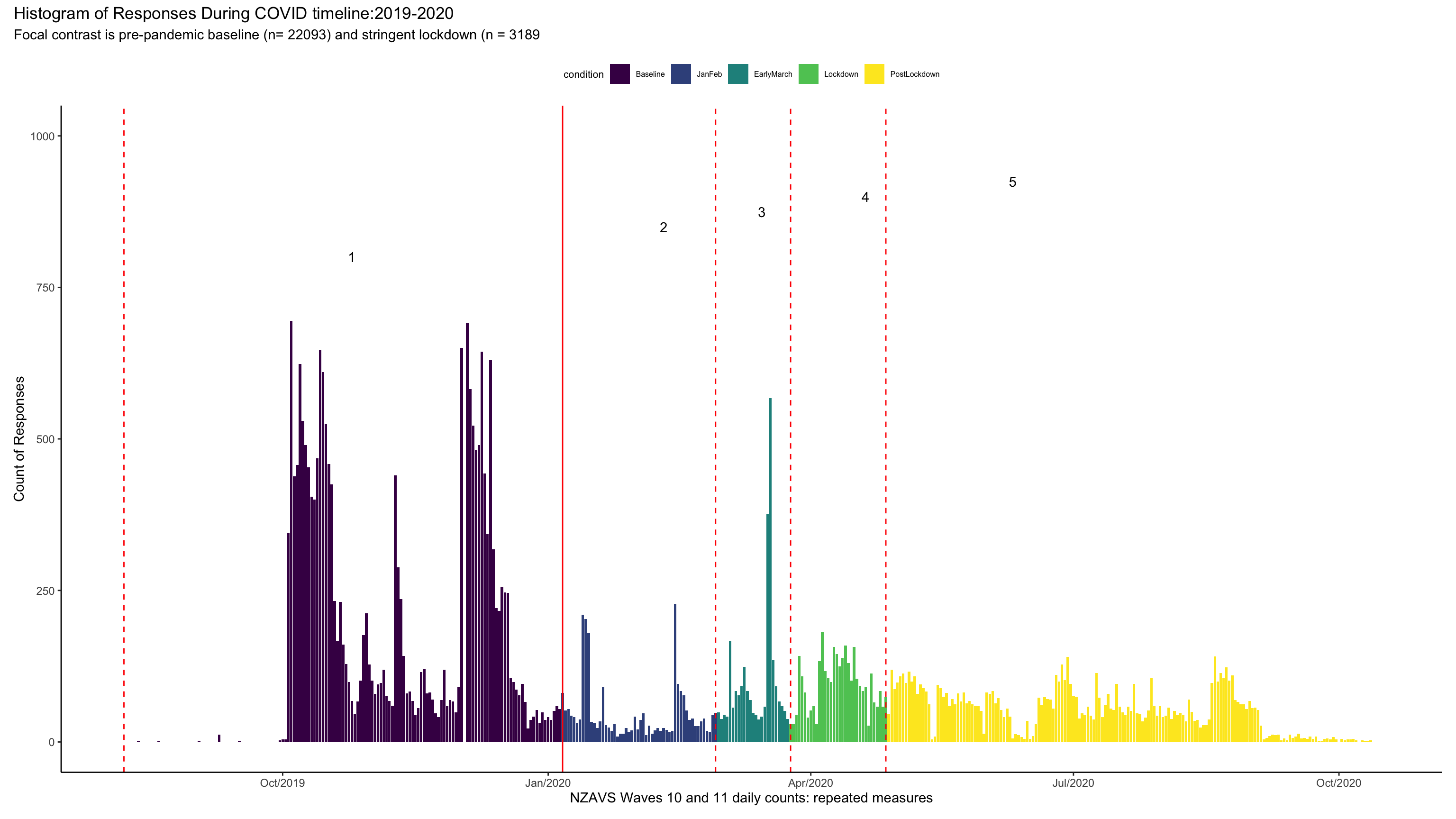
Appendix
Graph of timeline
Show code
# GRAPH
tline + plot_annotation(title = "Histogram of Responses During COVID timeline:2019-2020",
subtitle = "Focal contrast is pre-pandemic baseline (n= 22093) and stringent lockdown (n = 3189")